While the human body is rigid to a certain degree based on the skeleton, soft tissue is deformable in a way that does not conform to any rigid approximation, and most time soft tissues are often the target of medical interest. Predicting the deformation of soft tissue is a non-trivial task and thus a discipline of active research.
To make this problem more challenging, we want to predict deformation in ultrasound images and use it to create synthetic data to train other machine learning techniques for tasks such as segmentation, labeling, etc.
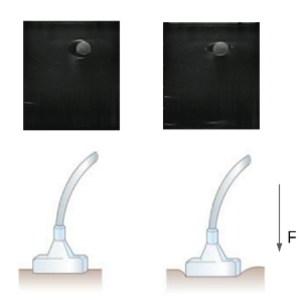
In our current dataset to train a segmentation model for segmenting vessels, we probe the subject with a fixed force, thus collecting our ultrasound images at one single force value. This biases our segmentation model to segment vessels at one force value. But in the case of high-tempo, traumatic scenarios on the battlefield, the ultrasound scanning might be done at higher/lower force values, thus deforming the vessels and thus failing the current segmentation model. To alleviate this problem, we propose to augment our dataset with synthetic ultrasound images at multiple locations of the subject at multiple ultrasound probe forces.
We plan to solve this problem by predicting the ultrasound image deformation/warping function which will be used to generate synthetic images. We will start by using B-Spline curves as the basis function to model the image warping function and use an iterative optimization problem, to find the location of the control points of the B-spline for the deformed image. Once we have the optimal B-spline curve as a function of the force applied, we will use that to create synthetic images at multiple force values and scanning locations. However, the true challenge of this work will lie in determining the true basis functions, beyond B-splines, that most accurately model the tissue deformation.
